-
 chevron_right
chevron_right
Scientists identify first known prehistoric person with Turner syndrome
news.movim.eu / ArsTechnica · Monday, 15 January - 22:37 · 1 minute
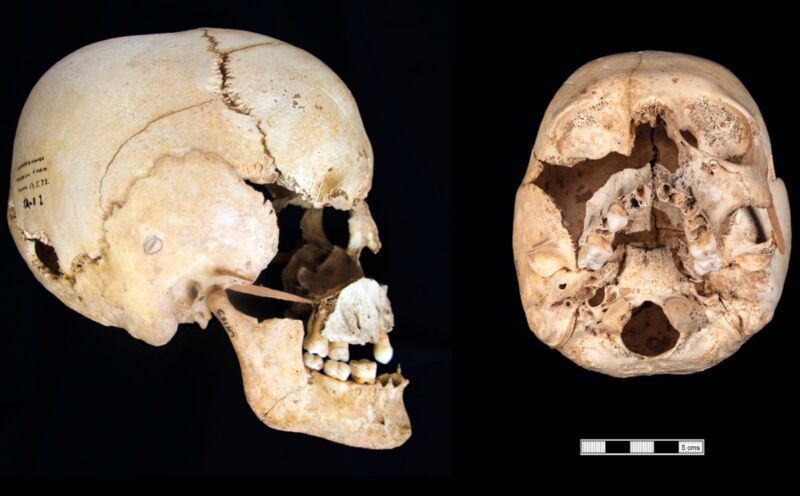
Enlarge / The cranium of an individual with mosaic Turner syndrome from an Iron Age site in Somerset, UK. (credit: K. Anastasiadou et al. 2024)
Turner syndrome is a genetic condition in which a (female) person has only one X chromosome instead of two. Scientists have used a new computational method for precisely measuring sex chromosomes to identify the first prehistoric person with this syndrome dating back some 2,500 years ago, according to a recent paper published in the journal Communications Biology. The team identified four other individuals with sex chromosomes outside the usual XX or XY designations: an early medieval individual with Jacobs syndrome (XYY) and three people from various periods with Klinefelter syndrome (XXY). They also identified an Iron Age infant with Down syndrome .
"It’s hard to see a full picture of how these individuals lived and interacted with their society, as they weren’t found with possessions or in unusual graves, but it can allow some insight into how perceptions of gender identity have evolved over time," said co-author Kakia Anastasiadou , a graduate student at the Francis Crick Institute.
Added co-author Rick Schulting, an archaeologist at the University of Oxford, “The results of this study open up exciting new possibilities for the study of sex in the past, moving beyond binary categories in a way that would be impossible without the advances being made in ancient DNA analysis."