Every person starts as just one fertilized egg. By adulthood, that single cell has turned into roughly 37 trillion cells, many of which keep dividing to create the same amount of fresh human cells every few months.
But those cells have a formidable challenge. The average dividing cell must copy—perfectly—3.2 billion base pairs of DNA, about once every 24 hours. The cell’s replication machinery does an amazing job of this, copying genetic material at a lickety-split pace of some 50 base pairs per second.
Still, that’s much too slow to duplicate the entirety of the human genome . If the cell’s copying machinery started at the tip of each of the 46 chromosomes at the same time, it would finish the longest chromosome—No. 1, at 249 million base pairs—in about two months.
“The way cells get around this, of course, is that they start replication in multiple spots,” says James Berger, a structural biologist at the Johns Hopkins University School of Medicine in Baltimore, who co-authored an article on DNA replication in eukaryotes in the 2021 Annual Review of Biochemistry. Yeast cells have hundreds of potential replication origins, as they’re called, and animals like mice and people have tens of thousands of them, sprinkled throughout their genomes.
“But that poses its own challenge,” says Berger, “which is, how do you know where to start, and how do you time everything?” Without precision control, some DNA might get copied twice, causing cellular pandemonium.
Keeping tight reins on the kickoff of DNA replication is particularly important to avoid that pandemonium. Today, researchers are making steps toward a full understanding of the molecular checks and balances that have evolved in order to ensure that each origin initiates DNA copying once and only once, to produce precisely one complete new genome.
Do it right, do it fast
Bad things can happen if replication doesn’t start correctly. For DNA to be copied, the DNA double helix must open up, and the resulting single strands—each of which serves as a template for building a new, second strand—are vulnerable to breakage. Or the process can get stuck. “You really want to resolve replication quickly,” says John Diffley, a biochemist at the Francis Crick Institute in London. Problems during DNA replication can cause the genome to become disorganized, which is often a key step on the route to cancer.
Some genetic diseases, too, result from problems with DNA replication . For example, Meier-Gorlin syndrome, which involves short stature, small ears, and small or no kneecaps, is caused by mutations in several genes that help to kick off the DNA replication process.
It takes a tightly coordinated dance involving dozens of proteins for the DNA-copying machinery to start replication at the right point in the cell’s life cycle. Researchers have a pretty good idea of which proteins do what, because they’ve managed to make DNA replication happen in cell-free biological mixtures in the lab. They’ve mimicked the first crucial steps in initiation of replication using proteins from yeast —the same kind used to make bread and beer—and they’ve mimicked much of the entire replication process using human versions of replication proteins , too.
The cell controls the start of DNA replication in a two-step process. The whole goal of the process is to control the actions of a crucial enzyme—called a helicase—that unwinds the DNA double helix in preparation for copying it. In the first step, inactive helicases are loaded onto the DNA at the origins, where replication starts. During the second step, the helicases are activated, to unwind the DNA.
Ready (load the helicase) …
Kicking off the process is a cluster of six proteins that sit down at the origins. Called ORC, this cluster is shaped like a double-layer ring with a handy notch that allows it to slide onto the DNA strands, Berger’s team has found.
In baker’s yeast, which is a favorite for scientists studying DNA replication, these start sites are easy to spot: They have a specific, 11- to 17-letter core DNA sequence, rich in adenine and thymine chemical bases. Scientists have watched as ORC grabs onto the DNA and then slides along, scanning for the origin sequence until it finds the right spot.
But in humans and other complex life forms, the start sites aren’t so clearly demarcated, and it’s not quite clear what makes the ORC settle down and grab on, says Alessandro Costa, a structural biologist at the Crick Institute who, with Diffley, wrote about DNA replication initiation in the 2022 Annual Review of Biochemistry. Replication seems more likely to start in places where the genome—normally tightly spooled around proteins called histones—has loosened up.
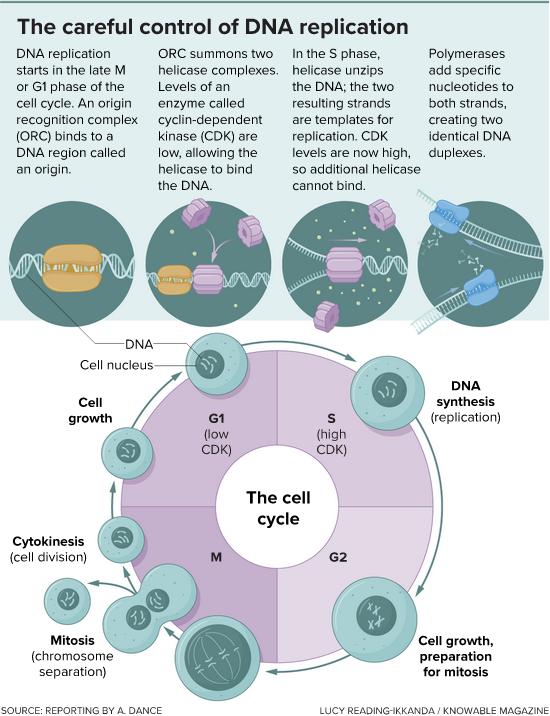
The initiation of DNA replication starts at the tail end of the previous cell division and continues through the cell cycle phase known as G1. DNA synthesis happens during the S phase. Levels of a protein called CDK are critical to ensuring that DNA is replicated once and only once. When CDK levels are low, helicases can jump onto the DNA and start to unwind it. But repeat binding does not happen because CDK levels rise, and this blocks the helicase from binding again. (credit: Knowable Magazine )
Once ORC has settled onto the DNA, it attracts a second protein complex: one that includes the helicase that will eventually unwind the DNA. Costa and colleagues used electron microscopy to work out how ORC lures in first one helicase, and then another . The helicases are also ring-shaped, and each one opens up to wrap around the double-stranded DNA. Then the two helicases close up again, facing toward each other on the DNA strands, like two beads on a string.
At first, they just sit there, like cars with no gas in the tank. They haven’t been activated yet, and for now the cell goes about its usual business.
Get set (activate the helicase)...
Things kick into high gear when a crucial molecule called CDK waves the green flag, jump-starting chemical steps that lure in even more proteins. One of them is DNA polymerase—what Costa calls the “typewriter” that will build new DNA strands—which hitches onto each helicase. Others activate the helicases, which can now burn energy to chug along the DNA.
As this occurs, the helicases change shape, pushing on one DNA strand and pulling on the other. This creates strain on the weak hydrogen bonds that normally hold the two strands together by the bases—the As, Cs, Ts and Gs that make up the rungs of the DNA ladder. The two strands get ripped apart. Costa and colleagues have observed how the two helicases untwist the DNA between them , and they’ve seen how the helicases keep the unbound bases stable and out of the way.
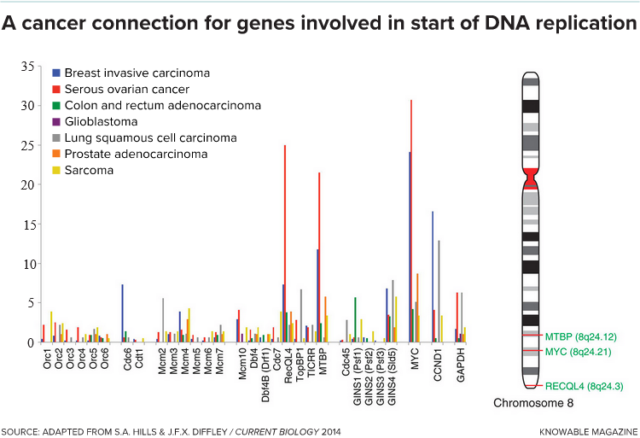
Left: Several genes involved in the initiation of DNA replication (horizontal axis) are amplified—that is, mistakenly copied in extra numbers — to varying degrees (vertical axis) in different cancers. Right: On chromosome 8, a cluster of three genes—shown in green text—are frequently amplified together in certain cancers. (credit: Knowable Magazine )
Go!
At first, both helicases are wrapped around both strands of DNA, and they can’t get very far like this, because they are facing each other and will just run into each other. But next, they each undergo a change in position, spitting one DNA strand or the other out of the ring. Now separated, they can jostle past each other, and replication proceeds apace.
Each helicase motors along its single strand, in the opposite direction from the other. They leave the origin behind and yank apart those hydrogen-bonded base pairs as they travel. The DNA polymerase is right behind, copying the DNA letters as they’re freed from their partners.
CDK’s second job is to stop any more helicases from hopping on the origins. Thus, there is one start of replication per origin, ensuring proper copying of the genome—although copying doesn’t begin at the same time at each site. The whole process of DNA replication, in human cells, takes about eight hours.
There is still plenty to be worked out. For one thing, the DNA that’s being copied is not a naked double helix. It’s wrapped around histones and attached to lots of other proteins that are busy turning genes on or off or making RNA copies of the genes . How do those jostling proteins affect each other and avoid getting in each other’s way?
Beyond this fascinating, fundamental biology—a remarkable process essential for all life on Earth—there are implications for diseases like cancer. Scientists already know that faulty replication can destabilize DNA, and an unstable genome that’s prone to mutation may be an early hallmark of cancer development. And they are further investigating links between replication proteins and cancer.
“I think that there are opportunities for therapeutic interventions for these systems,” says Berger, “once we have enough insights about how they work and what they look like.”
Amber Dance , a science writer in the Los Angeles area, also likes to break large tasks into smaller segments: It took her five days to complete the steps to draft this article. This article originally appeared in Knowable Magazine , an independent journalistic endeavor from Annual Reviews. Sign up for the newsletter .